JSOES Bongo Data Exploratory Analysis
Markus Min
2025-03-05
This page contains exploratory analysis for the Bongo net data from JSOES. Please note that because the Bongo net tows occur during daylight hours, the tow composition is biased towards taxa/life stages that do not vertically migrate. While different life stages of some species are counted separately for this data, for this exploratory analysis we summed abundances across life stages.
Data Overview
We will first explore the most common and most abundant taxa in this dataset.
| genus_species | common_name | mean_density_per_m3 | sd_density_per_m3 |
|---|---|---|---|
| Euphausiidae | Euphausiidae | 117.5 | 703.9 |
| Cirripedia | Barnacles | 34.8 | 757.1 |
| Chaetognatha | Chaetognatha | 3.5 | 16.1 |
| Euphausia pacifica | Euphausia Pacifica | 3.5 | 29.4 |
| Mitrocoma cellularia | Mitrocoma Cellularia (Cross Jellyfish) | 3.4 | 29.9 |
| Limacina | Limacina | 3.0 | 23.3 |
| 22 division fish egg | 22 Division Fish Egg | 2.9 | 8.6 |
| Engraulis mordax | Northern Anchovy | 2.5 | 25.1 |
| Calanus marshallae | Calanus Marshallae | 2.4 | 11.8 |
| Neotrypaea californiensis | Bay Ghost Shrimp | 2.4 | 159.8 |
| genus_species | common_name | prop_samples |
|---|---|---|
| Euphausia pacifica | Euphausia Pacifica | 0.83 |
| Euphausiidae | Euphausiidae | 0.80 |
| Thysanoessa spinifera | Thysanoessa Spinifera | 0.76 |
| Calanus marshallae | Calanus Marshallae | 0.70 |
| Cancer oregonensis/productus | Cancer Oregonensis/Productus | 0.70 |
| Chaetognatha | Chaetognatha | 0.70 |
| Themisto pacifica | Themisto Pacifica | 0.66 |
| Cirripedia | Barnacles | 0.64 |
| Limacina | Limacina | 0.49 |
| Crangonidae | Crangon | 0.46 |
Annual time series
This Shiny app can be used to explore the abundances of different taxa across the full length of the time series. In this Shiny app, I take the mean log density across the survey region to create a simple index of abundance.
Plotting distributions of some common taxa
I am currently developing a Shiny app that will allow you to visualize the distributions of different taxa. For now, I provide static maps of two focal taxa for this survey: Calanus marshallae (an abundant cold-water copepod) and Calanus pacificus (an abundant warm-water coepod). To demonstrate how we can visualize the abundance of these two key copepod species in space and time, we plot their density in survey catch below.
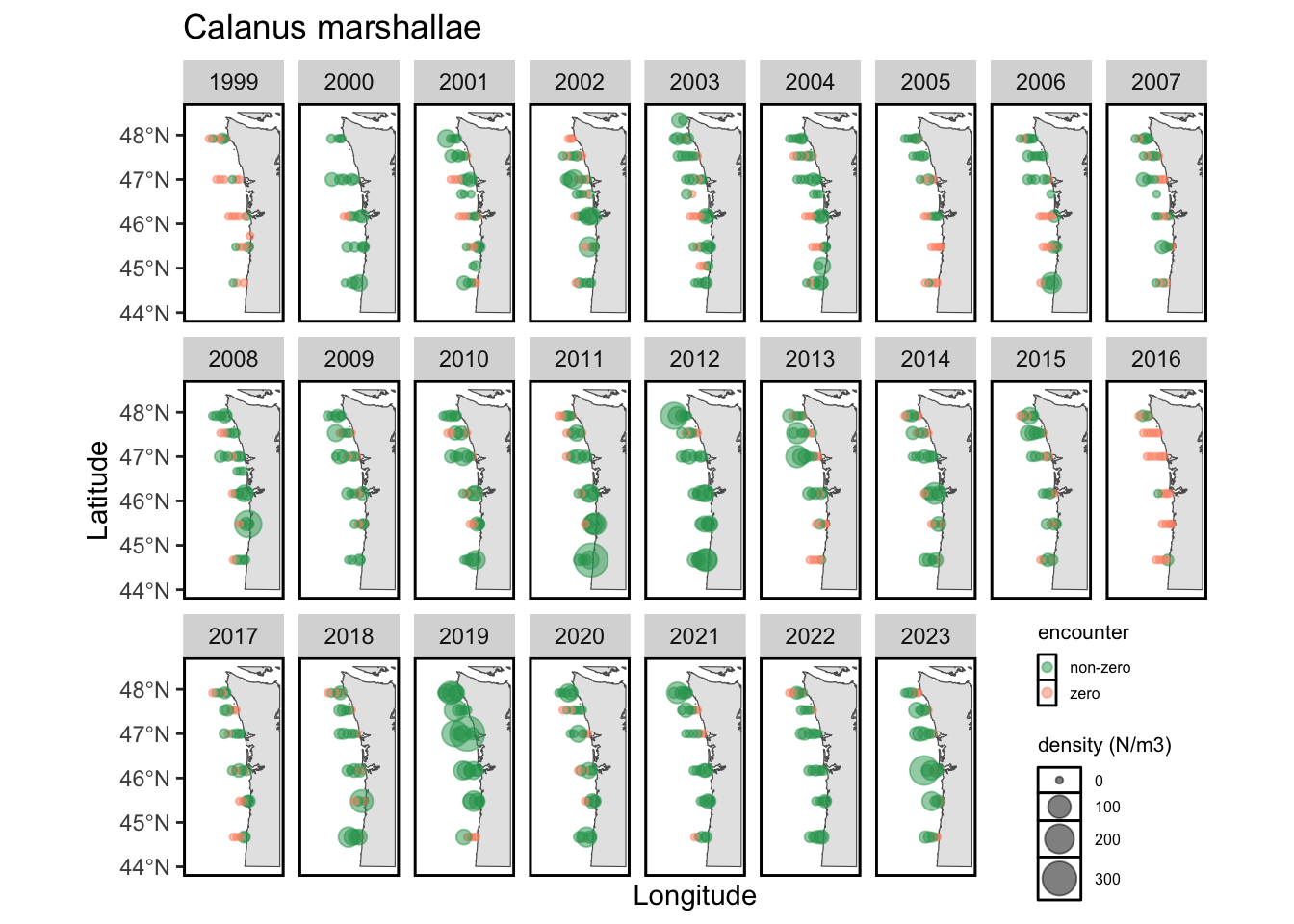
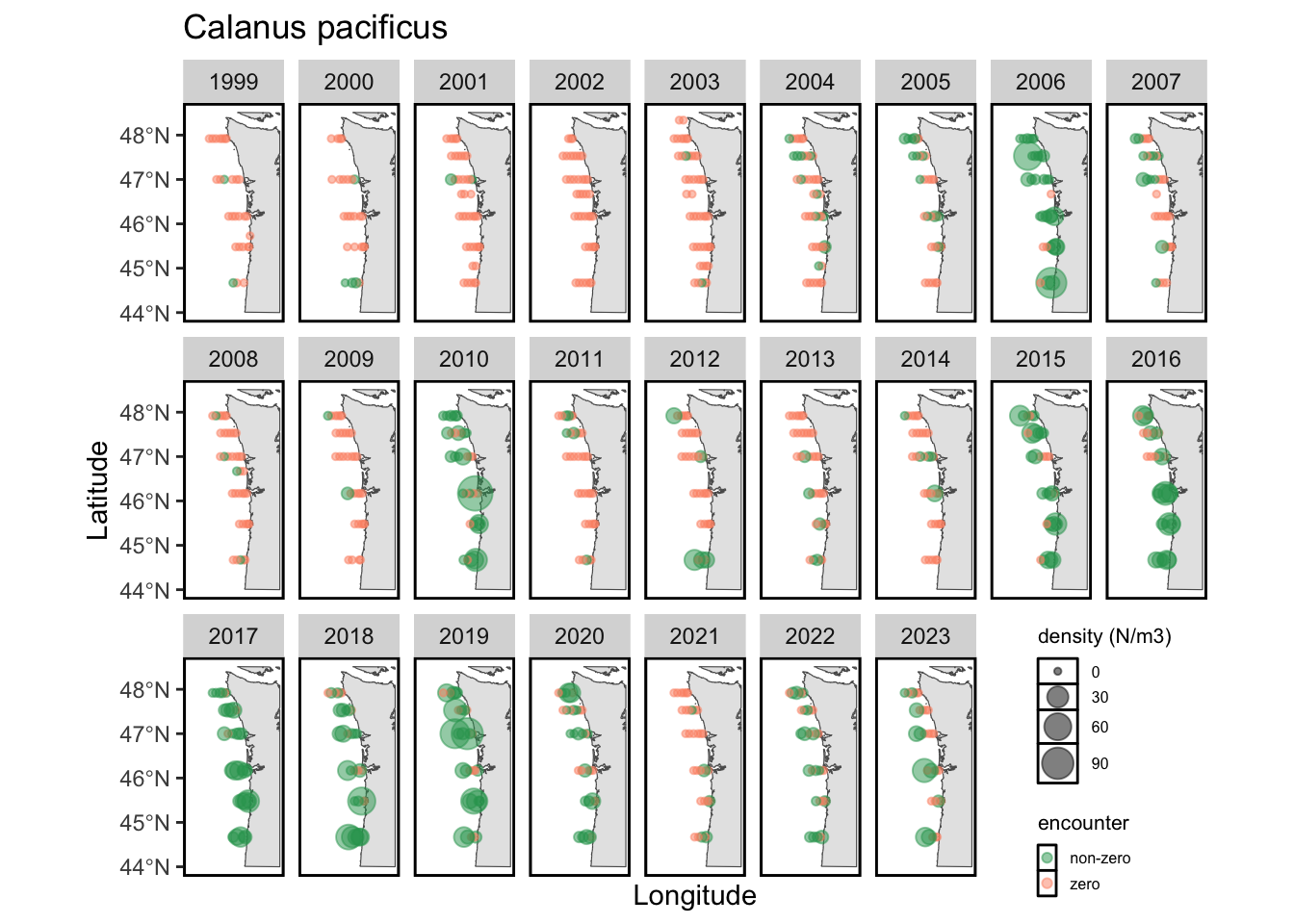
Temporal and Spatial Autocorrelation
Before fitting any spatiotemporal models, we must explore the spatial and temporal autocorrelation in the data.
Temporal structure
We can first inspect the autocorrelation in our Calanus marshallae and Calanus pacificus mean annual time series.
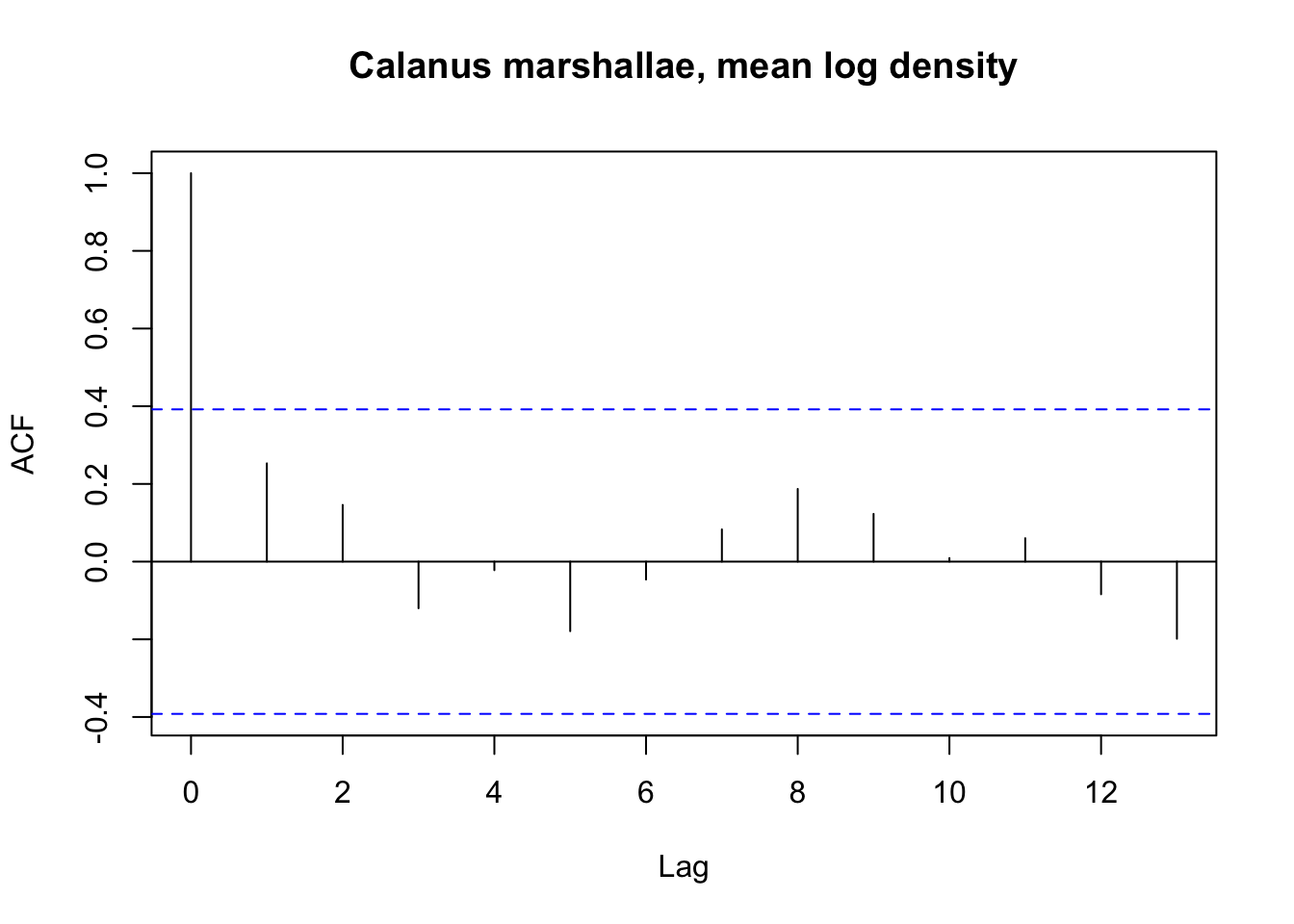
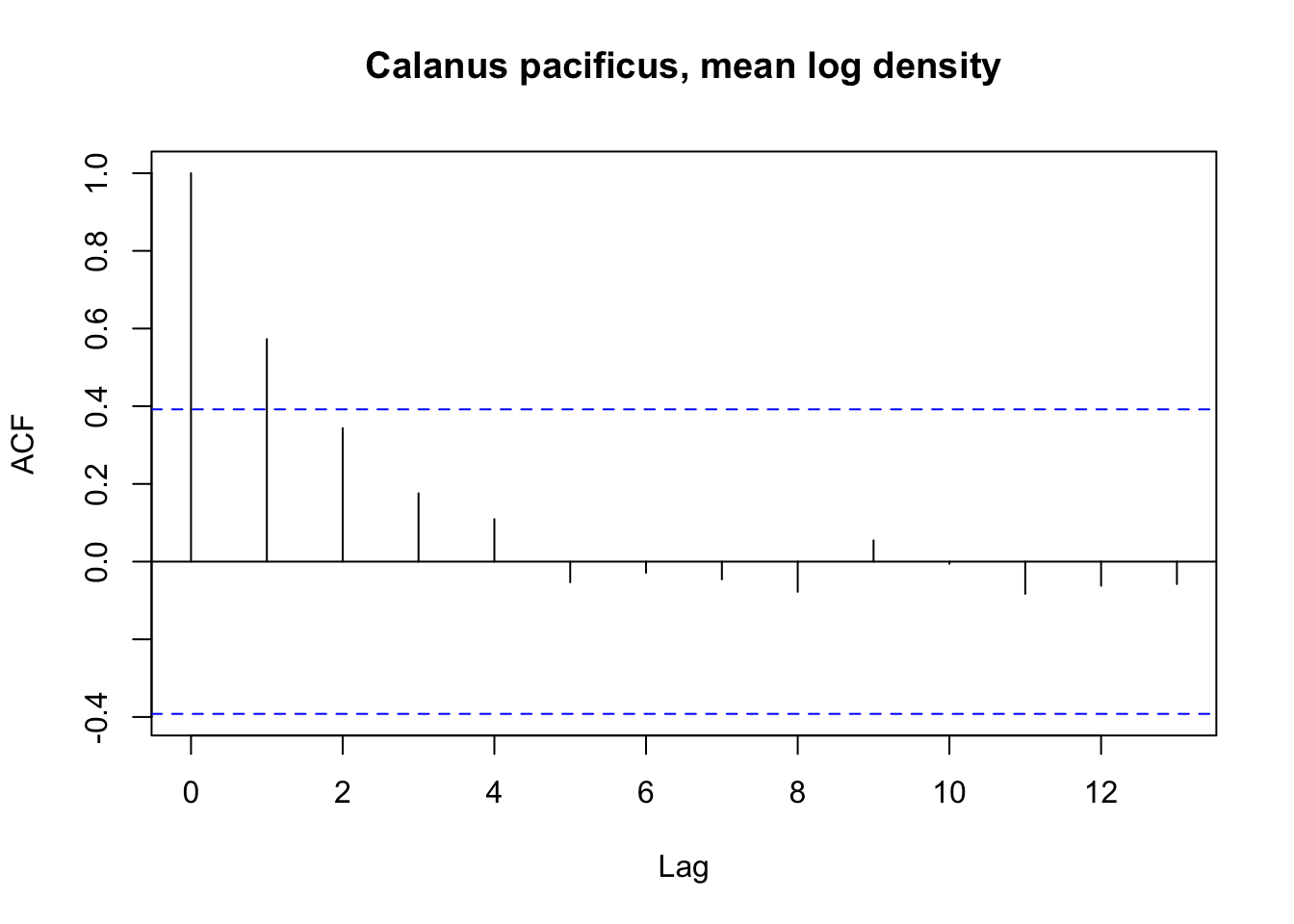
At the coastwide scale the Calanus marshallae time series does not show temporal autocorrelation, but the Calanus pacificus time series does at a lag of one year.
I also investigated temporal autocorrelation at the scale of individual stations, and found little evidence for temporal autocorrelation at this scale for these two taxa.
Spatial structure
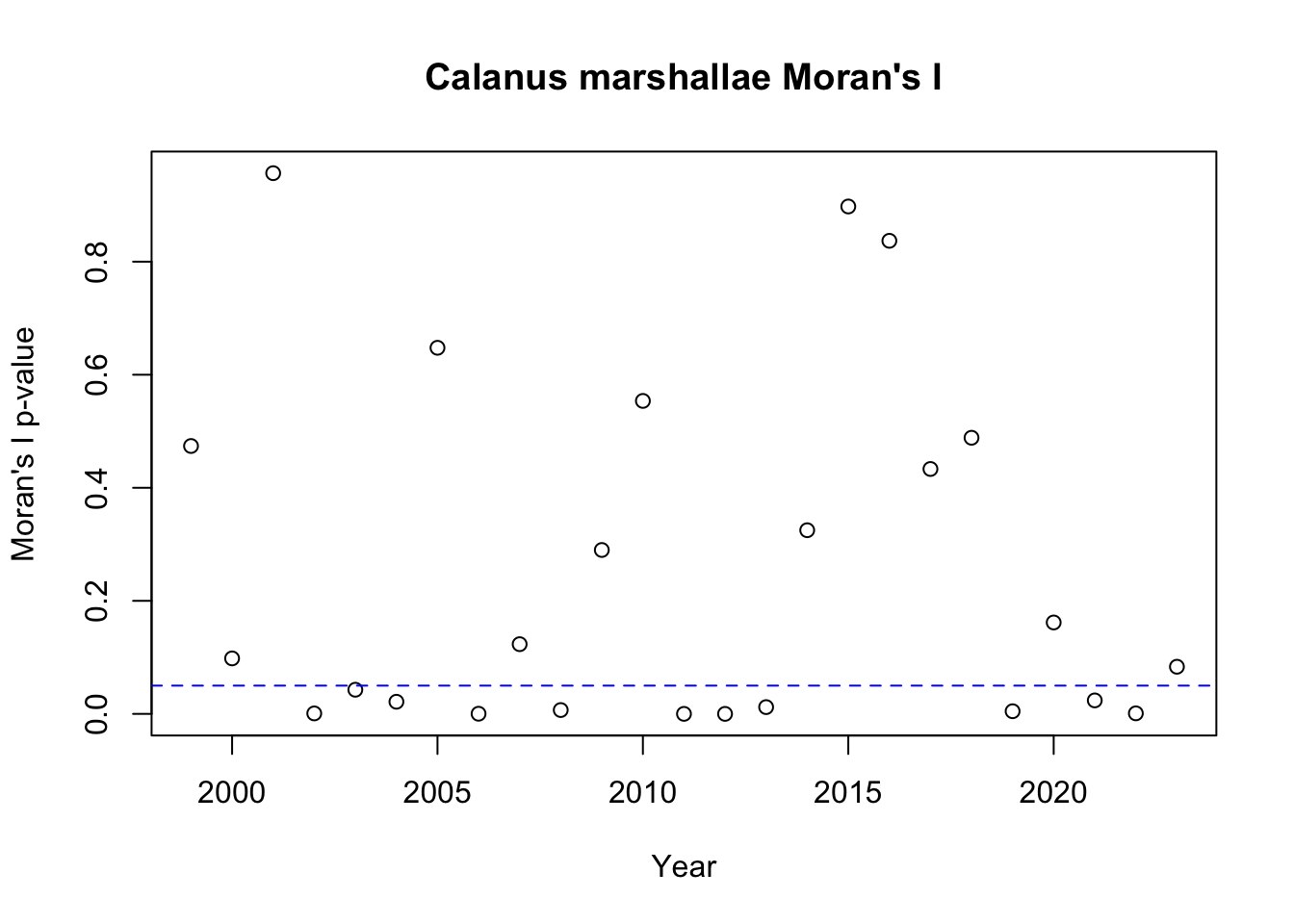
To investigate spatial autocorrelation, I calculated a metric known as Moran’s I. Moran’s I is a measure of the overall clustering of the spatial data and tests if there is support to reject the null hypothesis of no spatial structure. Given that we did not see much evidence for temporal autocorrelation, I calculated Moran’s I separately for each year. To summarize the Moran’s I results, I show the p-value for Moran’s I for each year, with the blue dashed line showing a p-value of 0.05.
We will first examine spatial autocorrelation in Calanus marshallae:
We will next examine spatial autocorrelation in Calanus pacificus:
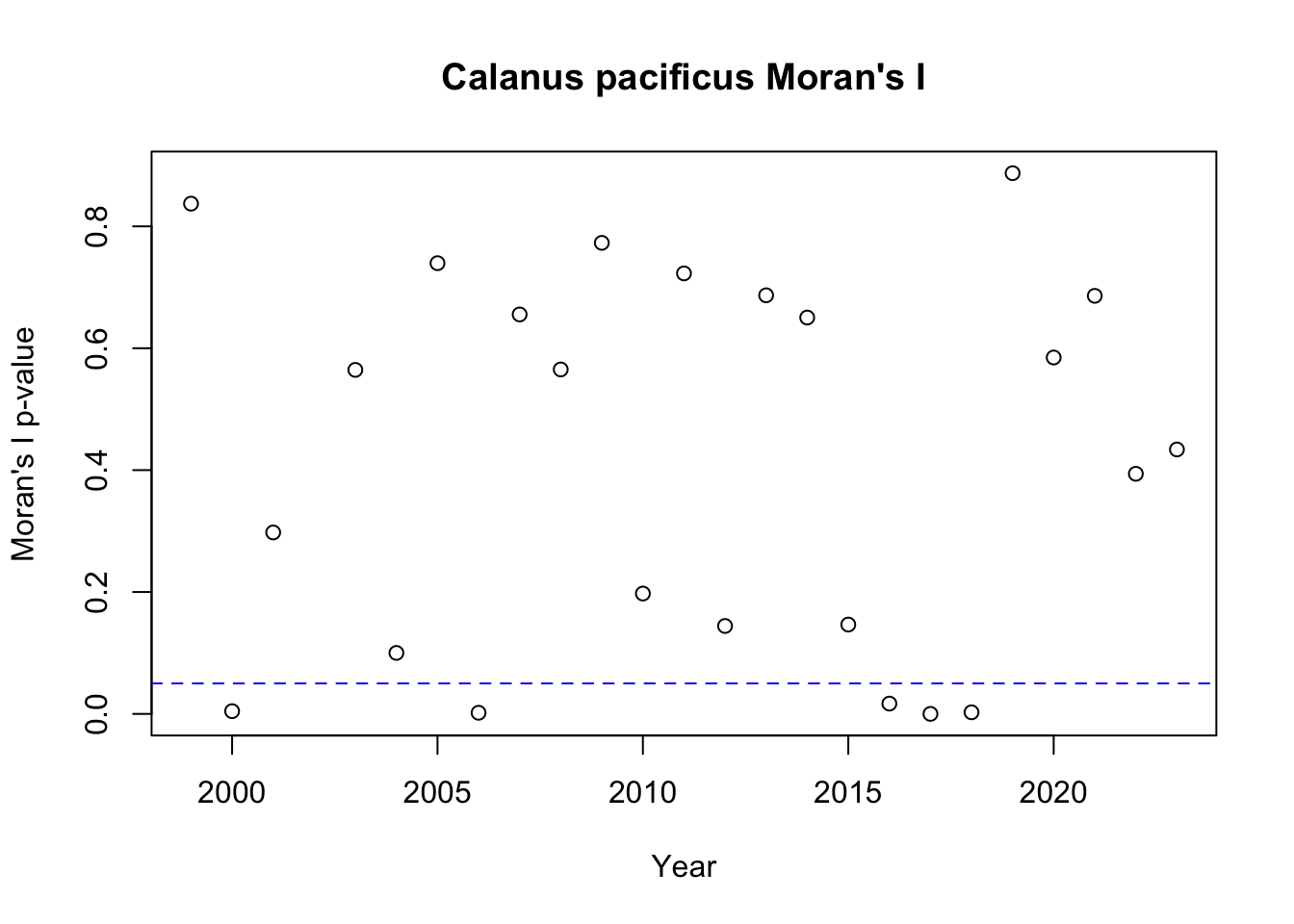
Based on the Moran’s I results, we see that there is evidence for spatial clustering in most years, especially for Calanus pacificus.